
This manuscript delineates the progression of microbial hydrobiology within the ambit of an ‘extramural’ university model, illustrated through the establishment of the Laboratory of Microbial Ecology (LEM) in Cantone Ticino and the Microbial Ecology Group at the University of Geneva. Situated within the premises of the Cantonal Institute of Microbiology, LEM has been instrumental in advancing scientific research and education at both graduate and undergraduate levels in the domains of clinical and environmental microbiology.
The core research at LEM concentrates on bacterial species prevalent in freshwater environments, particularly those with the potential for human contamination under specific circumstances, thereby underscoring their importance in clinical microbiology. The research also extends to microbial species crucial in biogeochemical cycles within both pristine and anthropogenically influenced freshwater ecosystems.
This manuscript offers a comprehensive overview of the scientific endeavors undertaken by LEM, detailing the studied bacterial species and mapping the evolution of analytical methodologies over the past three decades. Notably, advancements in microbiology and molecular biology have facilitated the identification and monitoring of emerging opportunistic pathogens such as Aeromonas, Yersinia, and Legionella in environmental settings, elucidating their transmission pathways from natural habitats to human infection.
A particular focus of the study is the permanently stratified ecosystem of Lake Cadagno, a meromictic lake adjacent to the Alpine Biology Centre. This lake serves as a paradigm for understanding biogeochemical processes in freshwater ecosystems, with an emphasis on the biological filter formed at the chemocline. This filter plays a pivotal role in sequestering toxic elements like sulfide, primarily through the activity of anaerobic key genera including Chromatium and Lamprocystis.
Moreover, the collaboration between the institutions in Geneva and Ticino has fostered numerous PhD research projects, the establishment of the Alpine Biology Center in Piora, and the development of an academic facility within the new premises of the Cantonal Institute of Microbiology in Bellinzona.
Copyright © 2023 E. Jack. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article pays homage to Prof. Gilbert Turian, who envisioned this possibility, by offering an overview of the scientific journey undertaken in the field of water microbiology.
The primary focus of the research has been on bacterial genera Aeromonas, Yersinia, Legionella, Acinetobacter, Chromatium, and Lamprocystis naturally inhabiting water. The investigation, conducted with increasingly sophisticated technical approaches, spans crucial stages that deserve reflection.
A series of works conducted at the Cantonal Institute of Microbiology in Ticino and the Microbial Ecology Laboratory of the University of Geneva revolve around the common theme of water-borne microorganisms [1]. To explore bacterial populations in various water niches and trace the pathways of contamination from the water environment to humans, a range of methods both phenotypic and molecular have been employed over the years.
These studies have not only contributed to the scientific understanding of water microbiology but have also resulted in numerous biology diplomas and doctoral theses presented at the University of Geneva.
The initial research endeavor in phage typing of Aeromonas bacteria marked a crucial step in characterizing strains isolated from both aquatic environments and clinical materials [2].The objective was to establish correlations between these two compartments. Various bacteriophages capable of lysing Aeromonas were isolated and employed to lyse strains within a concurrently established collection [3]. Additionally, “therapeutic” trials were conducted in fish breeding tanks (health baths) [4]. The outcomes underscored the existence of distinct phage types influenced by species and pathogenicity levels. However, certain strains remained non-typable using this criterion.
At that juncture, the official taxonomy of the Aeromonas genus acknowledged three mobile species (A. hydrophila, A. caviae, and A. sobria) and one immobile, psychrophilic species (A. salmonicida). Presently, the classification of Aeromonas has evolved, and taxonomic rearrangements, including the description of new species, subspecies, and biotypes, are underway. The genus, now subdivided into 17 genotypic species, of which only 14 are phenotypically recognizable, is currently part of a new family (Aeromonadaceae) (Fig. 1).
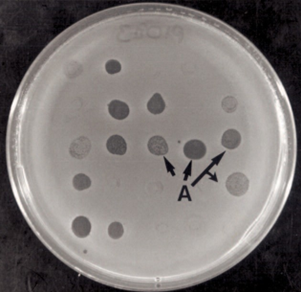
Figure 1: Bacterial MatAeromonas after 24 hours of incubation at 30°C. A: lysis zones caused by different bacteriophages.
Given the limitations encountered in employing phage typing for studying the epidemiology of water-borne pathogens, a novel approach called “Multilocus Enzyme Electroscopes” (MEE) was employed to characterize a collection of 120 strains of Aeromonas [5] [6]. The MEE method is grounded in assessing the electrophoretic mobility of metabolic enzymes, demonstrated on starch gels (see Figure 2). The migration distance is directly associated with the amino acid composition of the allelic form of the enzyme under scrutiny and, indirectly, with the nucleotide sequence of the corresponding gene allele. Through MEE, genetic diversity within the Aeromonas and Yersinia genera was revealed, highlighting the absence of epidemic clonal lineages and distinctly differentiating between strains of human origin and those of environmental origin refer to Figure 3.
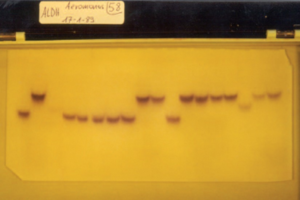
Figure 2: Specific staining of alanine dehydrogenase isoenzymes in lysates of different strains ofAeromonas after electrophoresis.
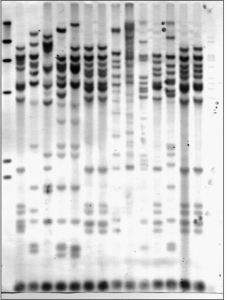
Figure 3: Ribotyping of strainsAeromonas
Taking an additional stride in microbial analysis, we employed “ribotyping,” a method involving the electrophoretic examination of bacterial DNA fragments, revealed using a probe derived from the ribosomal genes of E. coli. In the case of Aeromonas, this technique proved instrumental in achieving optimal discrimination between strains and facilitating their taxonomic classification. We applied “ribotyping” to scrutinize Aeromonas strains obtained from children exhibiting gastroenteric symptoms and their familial surroundings. Remarkably, identical ribotyping profiles surfaced in strains isolated from both patients and their environment. These findings underscored that, in certain instances, the origin of intestinal ailments in children could be directly linked to exposure to strains present in their family environment. Simultaneously, the widespread environmental presence of Aeromonas, coupled with their relatively low clinical incidence, hinted at the existence of strains with varying degrees of pathogenicity and host predisposition factors [7]. The study also delved into the immune response in the human intestine and explored proteins with toxic potential, including hemolysins ( Figure 4) [8].
Figure 4: Ribotyping of strainsAeromonas
Moreover, “ribotyping” yielded commendable results in characterizing Legionella strains (Gaia 1995) and Bacillus thuringiensis strains.

Figure 5: Phylogenetic tree obtained on the basis of rpoB gene sequences to differentiate species ofAeromonas.
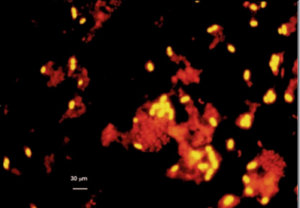
Figure 6: Chromatium okenii from the Cadagno Lake chemocline detected by “in situ” hybridization with a specific oligonucleotide probe labeled with Cy3
The development of probes specifically targeting these populations has subsequently enabled the exploration of their spatio-temporal variations over several years [10]. An insightful discovery emerged with the identification of clustered purple phototrophic bacteria belonging to the genus Rotsee, exhibiting diverse concentrations of sulfates and oxygen content [11]. This investigation significantly contributed to understanding the physical and chemical characteristics of the sediments, particularly regarding oxygen content and the presence of the two microbial groups considered.
Density Gradient Gel Electrophoresis (DGGE) found application in comparing bacterial populations within activated sludge from wastewater treatment plants. Additionally, it played a role in evaluating the impact of antibiotic residue constraints on these populations.
The combined use of PCR and gene sequencing further enhanced the characterization of Aeromonas popoffii species. A noteworthy aspect involved the association of Lamprocystis with sulfate-reducing bacteria of the genus Desulfocapsa (Figure 7). Pure culture studies conducted during a doctoral thesis [12] expanded the research scope into the field of physiology. DGGE, in conjunction with phylogeny and FISH, was subsequently employed to comparatively study sulfate-reducing bacteria and methanogenic Archaea in the sediments of Lakes Cadagno.
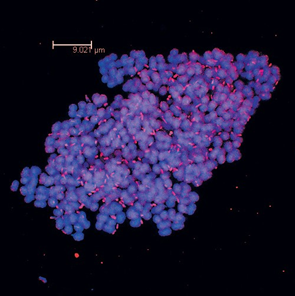
Figure 7: Image taken with a confocal microscope. Clusters of phototrophic bacteria belonging to the genusLamprocystis (cells/cocci) with sulfate-reducing bacteria of the genus Desulfocapsa (rod cells).
Advancements and Continued Exploration.The genus Aeromonas, known for its intricate taxonomy posing challenges to epidemiological studies, has been subject to insightful investigations employing robust tools. These methods have not only aided in overcoming taxonomic hurdles but also facilitated the differentiation of the three genomic species exhibiting the Yersinia frederiksenii phenotype. Furthermore, they have played a crucial role in characterizing a strain of Pseudomonas [13].
The experience gained from these tools has paved the way for ongoing studies within the broader realm of microorganisms inhabiting water habitats. Molecular techniques, in particular, continue to be instrumental in delving deeper into the expression of noteworthy and biologically significant phenotypes. This includes the exploration of RubisCO activity, the investigation of toxic protein presence and secretion by bacteria, and the scrutiny of antibiotic resistance. As research persists along these lines, these powerful tools promise to unravel further insights into the dynamic world of microorganisms in aquatic environments.